Doubling the conventional resolution limit for live cell time-lapse imaging.
The N-SIM S Super Resolution Microscope is a unique high-speed structured illumination system that achieves acquisition speeds of up to 15 fps, enabling fast biological processes to be captured at twice the spatial resolution of conventional light microscopes (up to 115nm in XY). Combining the N-SIM S and a confocal microscope gives you the flexibility to select a location in the confocal image and switch to super-resolution to view the desired part of the location in minute detail.

Key Features
High-speed super-resolution imaging at 15 fps
Nikon’s new high-speed structured illumination system utilizes a novel pattern modulation technology to generate fast and precise switching of illumination patterns. The N-SIM S achieves incredible acquisition speeds (up to 15 fps*), enabling super-resolution time-lapse imaging of live cells and intracellular dynamics.
* 2D-SIM mode, 512 x 512 pixels, 2 msec exposure time
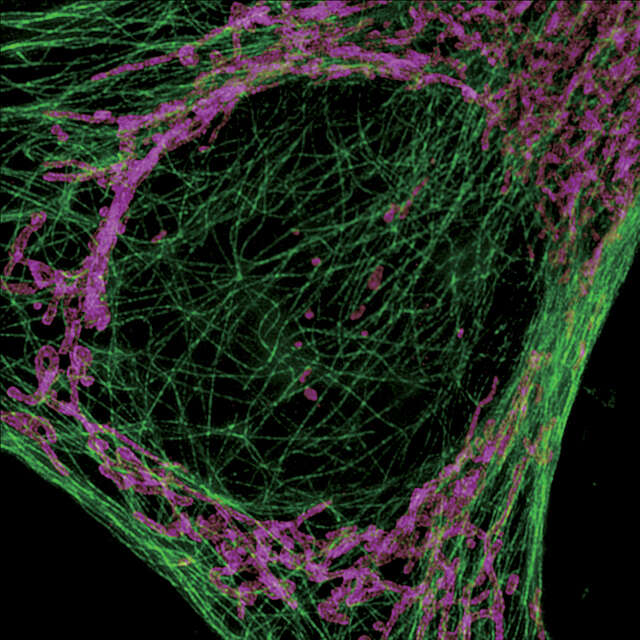
Endosomes of a COS7 cell labeled with YFP. Rapid movement of endosomes is captured at high resolution. This video shows a comparison with a widefield image. Image acquisition speed: 6 fps Imaging mode: 3D-SIM Image courtesy of: Yasushi Okada, M.D., Ph.D., Department of Physics, Graduate School of Science, The University of Tokyo
Growth cone of NG108 cell labeled with GFP-Lifeact for F-actin. Formation of an actin mesh is captured at high-speed. This video shows a comparison with a widefield image.
Image acquisition speed: 10 fps
Imaging mode: TIRF-SIM
Image courtesy of: Drs. Minami Tanaka and Kaoru Katoh, The National Institute of Advanced Industrial Science and Technology (AIST)
Histone H2B-GFP expressing HeLa cell. Visualization of fine movements of chromatin domains in different locations. This video shows a comparison with a widefield image.
Image acquisition speed: 3.9 fps
Imaging mode: 3D-SIM
Image courtesy of: Yuko Sato, Ph.D. and Hiroshi Kimura, Ph.D., Cell Biology Center, Institute of Innovative Research, Tokyo Institute of Technology
ER of COS7 cell labelled with SGFP2-sec61b. Fine movements of endoplasmic reticula can be captured and visualized with reconstruction. This video shows a comparison with a widefield image.
Image acquisition speed: 3.9 fps
Imaging mode: 3D-SIM
Image courtesy of: Ikuo Wada, Ph.D., Department of Cell Science, Institute of Biomedical Science, Fukushima Medical University, Drs. Shizuha Ishiyama and Kaoru Katoh, The National Institute of Advanced Industrial Science and Technology (AIST)
Live-cell imaging at double the resolution of conventional light microscopes
The N-SIM S utilizes Nikon's innovative approach to Structured Illumination Microscopy technology. By pairing this powerful technology with Nikon’s renowned objectives, which achieve an unparalleled numerical aperture of 1.49, the N-SIM S nearly doubles (to approximately 115 nm*) the spatial resolution of conventional light microscopes, enabling detailed visualization of minute intracellular structures and their interactions.
* This value is the FWHM measurement of 100 nm beads excited with a 488 nm laser in 3D-SIM mode. In TIRF-SIM mode, 86 nm is achieved using 40 nm beads excited with a 488 nm laser.
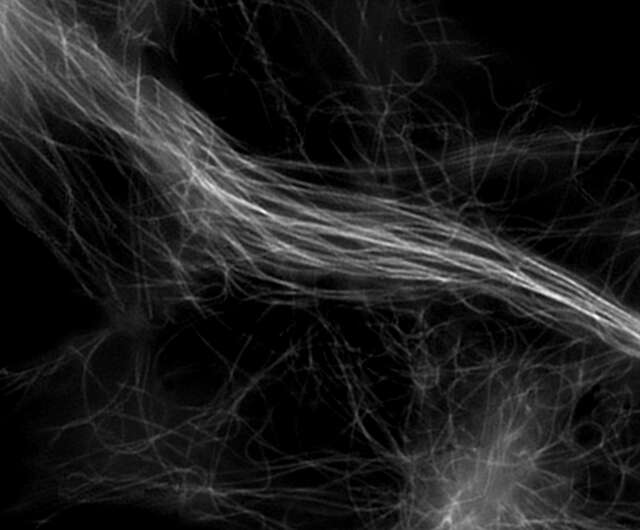
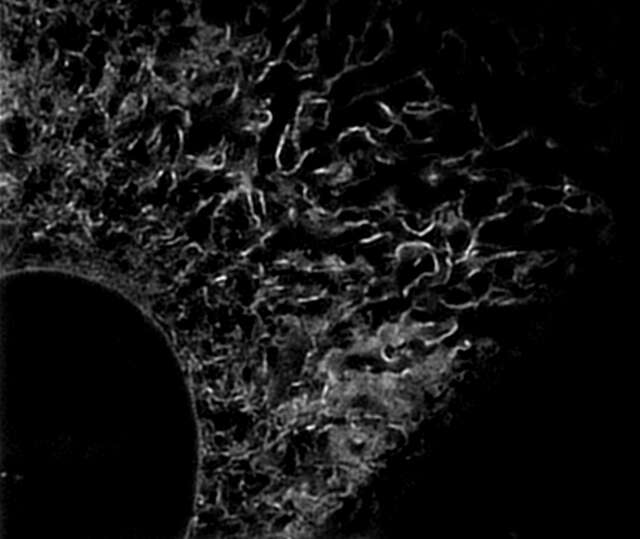
Super-resolution image (3D-SIM)
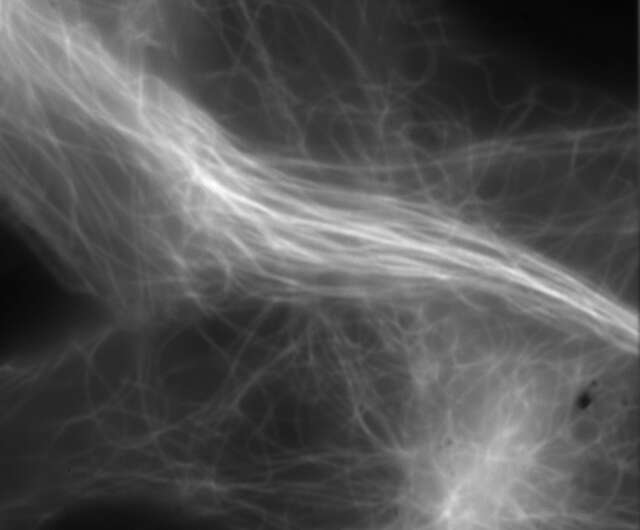
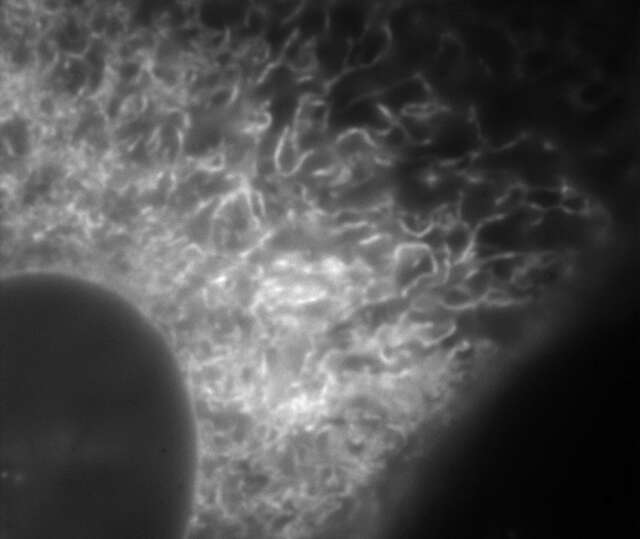
Conventional widefield image
Microtubules in B16 melanoma cell labeled with YFP
Objective: CFI Apochromat TIRF 100XC Oil (NA 1.49)
Image capturing speed: approximately 1.8 sec/frame (movie)
Reconstruction method: Slice
Photographed with the cooperation of: Dr. Yasushi Okada, Laboratory for Cell Polarity Regulation, Quantitative Biology Center, RIKEN
Super-resolution image (3D-SIM)
Conventional widefield image
Endoplasmic reticulum (ER) in living HeLa cell labeled with GFP
Objective: CFI Apochromat TIRF 100XC Oil (NA 1.49)
Image capturing speed: approximately 1.5 sec/frame (movie)
Reconstruction method: Slice
Photographed with the cooperation of: Dr. Ikuo Wada, Institute of Biomedical Sciences, Fukushima Medical University School of Medicine
Automatic switching between illumination modes
The newly-developed high-speed structured illumination technology enables fast acquisition rates, automatic switching between illumination modes, and automated optimization of structured illumination patterns for different wavelengths and magnifications. This expanded automation enables fast 2-color TIRF-SIM imaging, as well as multiplexing of different SIM modalities. The N-SIM S provides easy-to-use, streamlined workflows, whether it be for single-mode or multi-modal imaging experiments.
Acquire larger fields of view
N-SIM S can acquire super-resolution images with a large field of view of 66 µm square. This larger imaging area enables very high throughput for applications/samples that benefit from larger fields of view, such as a neurons, reducing the amount of time and effort required to obtain data.
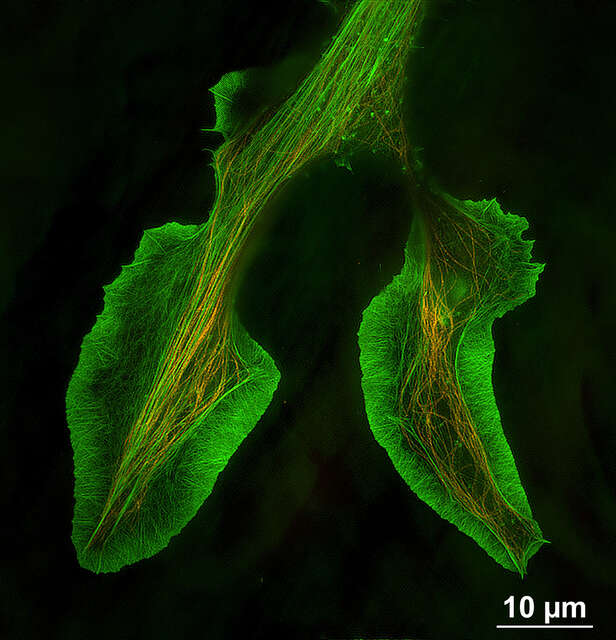
Two-color TIRF-SIM imaging of growth cone of NG108 cell labeled with Alexa Fluor® 488 for F-actin (green) and Alexa Fluor® 555 for microtubules (orange)
Reconstructed image size: 2048 x 2048 pixels (66 μm x 66 μm with a 100X objective)
Sample courtesy of: Drs. Shizuha Ishiyama and Kaoru Katoh, The National Institute of Advanced Industrial Science and Technology (AIST)
Various observation modes
TIRF-SIM/2D-SIM mode
This mode captures super-resolution 2D images at high speed with incredible contrast. The TIRF-SIM mode enables Total Internal Reflection Fluorescence observation at double the resolution of conventional TIRF microscopes, facilitating a greater understanding of molecular interactions at the cell surface.
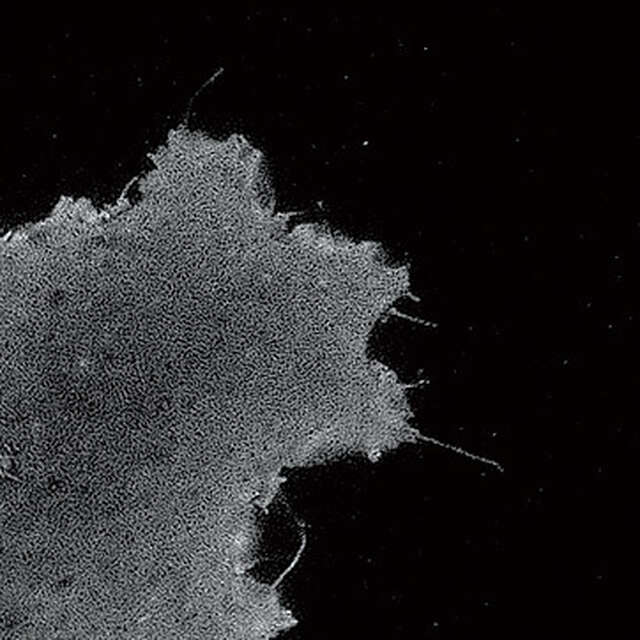
TIRF-SIM image
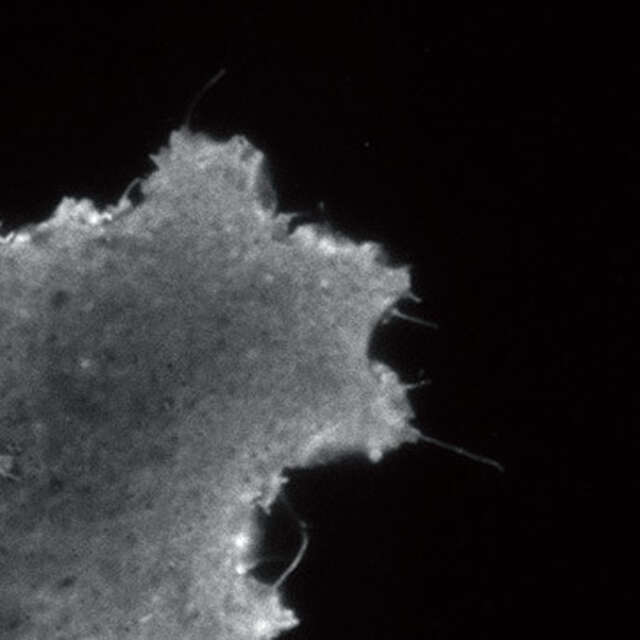
Conventional TIRF image
Plasma membrane of B16 melanoma cell labeled with YFP
Objective: CFI Apochromat TIRF 100XC Oil (NA 1.49)
Photographed with the cooperation of: Dr. Yasushi Okada, Laboratory for Cell Polarity Regulation, Quantitative Biology Center, RIKEN
3D-SIM mode
The 3D-SIM mode generates structured illumination patterns in three dimensions to deliver a two-fold improvement in lateral and axial resolutions. Two reconstruction methods (“slice” and “stack”) are available to optimize results according to application requirements (e.g. sample thickness, speed, etc.). Slice reconstruction is suitable for imaging living cells at specific depths, as it allows axial super-resolution imaging with optical sectioning at 300 nm resolution. Stack reconstruction, based on Gustafsson’s theory, is suitable for acquisition of volume data as it can image thicker specimens with higher contrast than slice reconstruction.
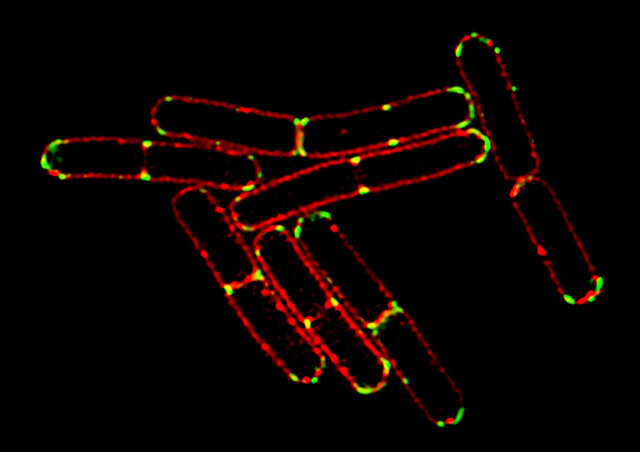
3D-SIM image
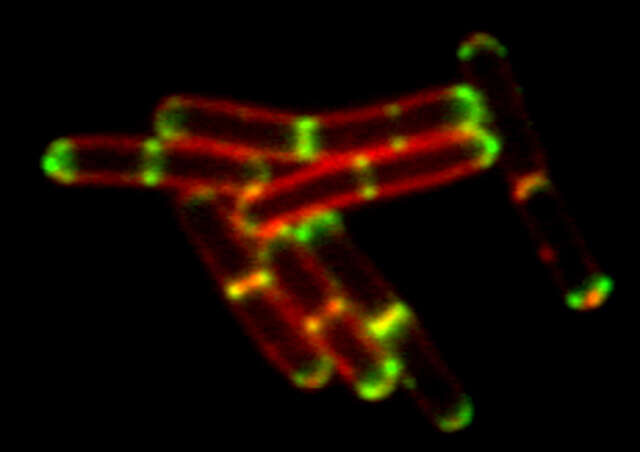
Conventional widefield image
Bacillus subtilis bacterium stained with membrane dye Nile Red (red), and expressing the cell division protein DivIVA fused to GFP (green). The super-resolution microscope enables accurate localization of the protein during division.
Reconstruction method: Slice
Images courtesy of: Drs. Henrik Strahl and Leendert Hamoen, Centre for Bacterial Cell Biology, Newcastle University
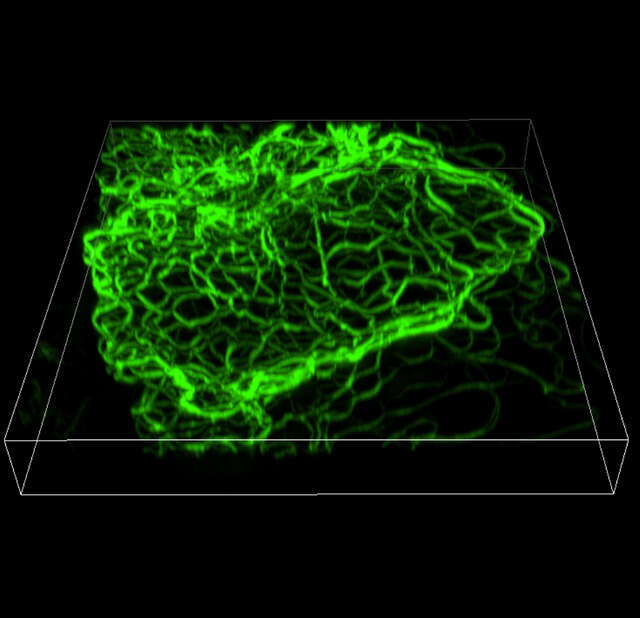
3D-SIM (Volume view)
Width: 26.16 μm, Height: 27.11 μm, Depth: 3.36 μm
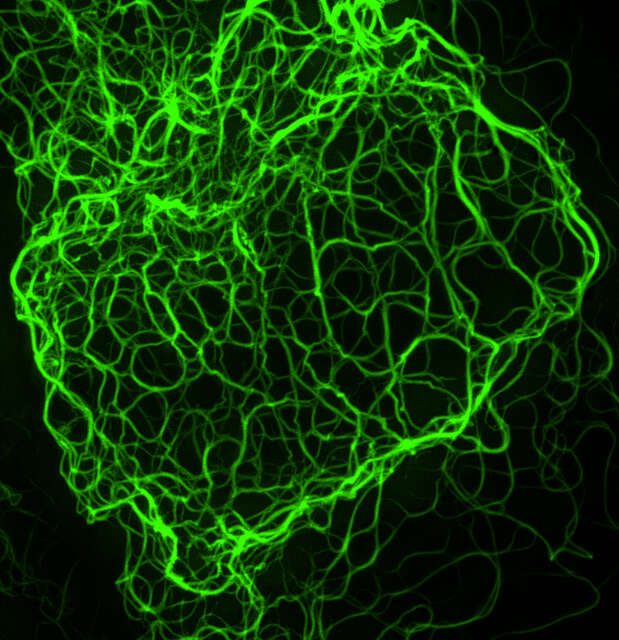
3D-SIM (Maximum projection)
Mouse keratinocyte indirectly immunolabeled for keratin intermediate filaments and visualized with Alexa Fluor® 488 conjugated secondary antibodies.
Reconstruction method: Stack
Image courtesy of: Dr. Reinhard Windoffer, RWTH Aachen University
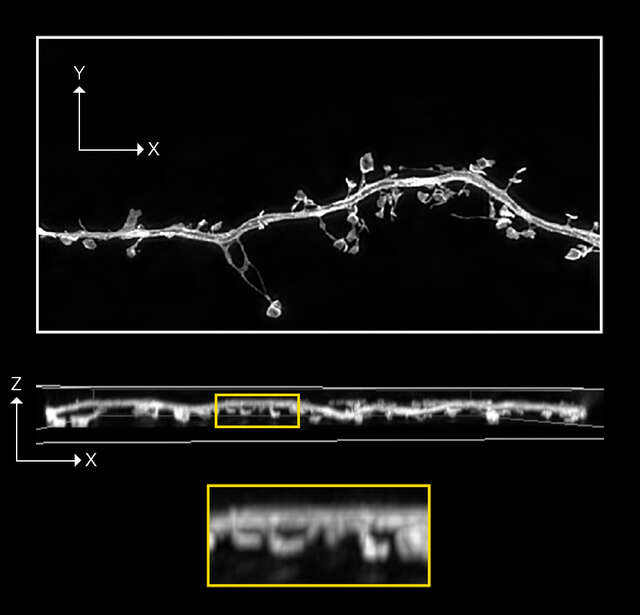
N-SIM S image
Z-stack of 3D-SIM ,19 stps, Z range: 2 μm
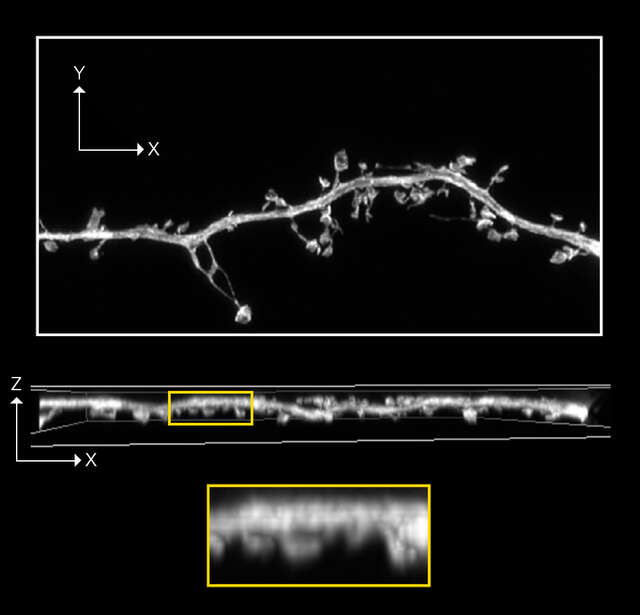
Confocal A1R image with 0.4AU Deconvolution
Z-stack ,19 stps, Z range: 2 μm
Sample information: Dendritic spine in mouse hippocampal neuron expressing GFP
Image courtesy of: Drs. Yutaro Kashiwagi and Shigeo Okabe, Department of Cellular Neurobiology, Graduate School of Medicine and Faculty of Medicine, The University of Tokyo.
Quantitative spine analysis with Z-stack of 3D-SIM, 35 stps, Z range: 4.2 um
Exposure time 100 msec, 120 sec interval
Time-lapse of 11 times
Excitation wavelength 488 nm
Sample information: Dendritic spine in mouse hippocampal neuron expressing GFP
Movie courtesy of: Drs. Yutaro Kashiwagi and Shigeo Okabe, Department of Cellular Neurobiology, Graduate School of Medicine and Faculty of Medicine, The University of Tokyo.
Simultaneous two-channel imaging
Simultaneous two-color imaging is possible by utilizing an optional Two Camera Imaging Adaptor* and two sCMOS cameras.
* Hamamatsu Photonics K.K.
| Immediately before drug application | 10 minutes after |
20 minutes after | 30 minutes after |
Growth cone of NG108 cells. Fascin (green): GFP-Fascin, Actin (red): LifeAct-KO Objective Lens : CFI SR HP Apochromat TIRF 100xC oil
Using image splitting optical system, dual color time-lapse images were captured after application of drug solution. The time course was recorded that fascin was dissociated from the actin bundles and filamentous actin changed their pattern. Co-localization of fascin and actin and dissociation of fascin were well demonstrated. Optic aberrations and image distortion were adjusted to be less than 30 nm.
Movie courtesy of: Dr. Minami Tanaka, Dr. Kaoru Katoh, Biomedical Research Institute Molecular Neurobiology Group, National Institute of Advanced Industrial Science and Technology
Seamless switching between imaging modalities for multi-scale experiments
The N-SIM S can be simultaneously combined with a confocal microscope such as the AX/AX R. A desired location in a sample can be specified in a low-magnification/large FOV confocal image and acquired in super-resolution by simply switching the imaging method. Combining a confocal microscope with a super-resolution system can provide a method for gaining larger contextual views of super-resolution information.
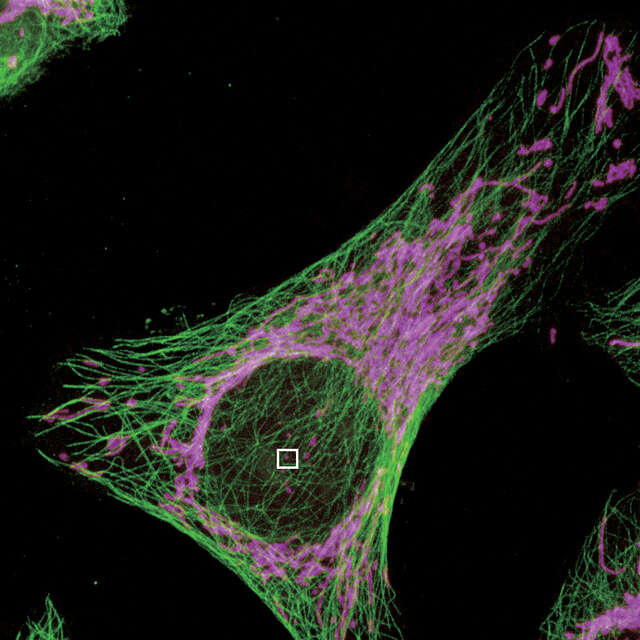
Select the location to acquire a SIM image in a confocal image
Acquire the SIM image of the selected location
Objectives for super-resolution microscopes
Silicone immersion objectives
Silicone immersion objectives use high viscosity silicone oil with a refractive index close to that of live cells as an immersion liquid. Because of this improved refractive index compatibility, these objectives can provide improved photon collection capability and resolution when performing super-resolution imaging deeper into the specimen. They exhibit superior chromatic aberration correction and high transmittance over a broad range of wavelengths.

CFI SR HP Plan Apochromat Lambda S 100XC Sil
Mouse brain section labeled with tdTomato expressing neurons
Immersion objectives
SR series objectives are aligned and inspected using wavefront aberration measurement technology to ensure the lowest possible asymmetric aberration and superior optical performance required for super-resolution imaging. HP model objectives provide ultra-high power laser excitation durability and improved axial chromatic aberration correction, eliminating the need to switch objectives between the N-SIM S and N-STORM systems. The AC-type objectives that support the Auto Correction Collar of the Ti2-E microscope enable precise and easy adjustment of the correction collar.

CFI SR HP Apochromat TIRF 100XC Oil

CFI SR Plan Apochromat IR 60XC WI

CFI SR HP Apochromat TIRF 100XAC Oil
Dry objectives
The N-SIM S is compatible with dry objectives, making both super-resolution imaging and confocal imaging available without switching lenses. Low-magnification, wide field-of-view dry lenses enable high resolution observation even at the periphery of sample tissues.
* Dry objectives support 2D-SIM and 3D-SIM (slice reconstruction)